Using optical tweezer technology, Technion researchers were able to gain a greater understanding of the poorly understood DNA packaging process, which impacts how genes are expressed.
Article published at www.jpost.com on July 6, 2021.

Scientists from the Technion-Israel Institute of Technology have used “laser tweezers” to understand the structure of DNA better than ever before, shedding light on poorly understood mechanisms that influence how genes express themselves in the human body.
Chromatin is found in DNA, the essential code in the human body that provides instructions needed for function and development. Though there is said to be around two meters total of DNA in the human body, found in the nucleus of every single cell, it is compressed into just tens of microns in size. This is because of how DNA itself is packaged, formed into a compact structure known as chromatin.
Chromatin itself is organized by wrapping the DNA strands around a specific protein called histones. The spool-like structure it eventually forms is said to resemble beads on a string. These “strings” are then connected with a special type of histone called a linker histone, which helps the strands form into more complex structures called chromatosomes.
The advantage of packaging the human genome in this form is that it makes it possible for it to actually physically fit within the cell. However, it makes accessing it more difficult, which can pose problems for mechanisms within the cell that are supposed to read the DNA.
The result is that the way a gene is ultimately expressed becomes dependent on a particular method of packaging. How exactly this works is still a mystery, and scientists have struggled to find an answer. But one thing that has been uncovered is the role of linker histones in the organization of this packaging. In other words, if a linker histone malfunctions, it could lead to improper packaging, which can result in non-ideal gene expression.
According to some experts, it is believed that the end result of linker histone malfunctions could manifest as autism or serious diseases like cancer. But how the linker histones actually bind DNA in the first place remains a mystery, making it even more difficult to investigate the issue using conventional methods.
But an unconventional method is exactly what Dr. Sergei Rudnizky used. The scientist and his team developed a new method based on “optical tweezers,” a method that uses a focused laser beam to capture individual molecules and exert force on them.
The method itself was pioneered by Jewish-American scientist Arthur Ashkin in 1986. It was based on years of research he had conducted in the 1970s, which had later formed the basis for the 1986 work by Steven Chu on using optical tweezing on cooling and trapping neural atoms. Chu would win the Nobel Prize in Physics for this in 1997, and would later go on to serve as US secretary of energy from 2009 to 2013. Ashkin himself would later win the Nobel Prize in Physics for it in 2018.
The technology was also later employed in other sectors, and in 2010 was adopted by Tel Aviv University for nanotechnology research.
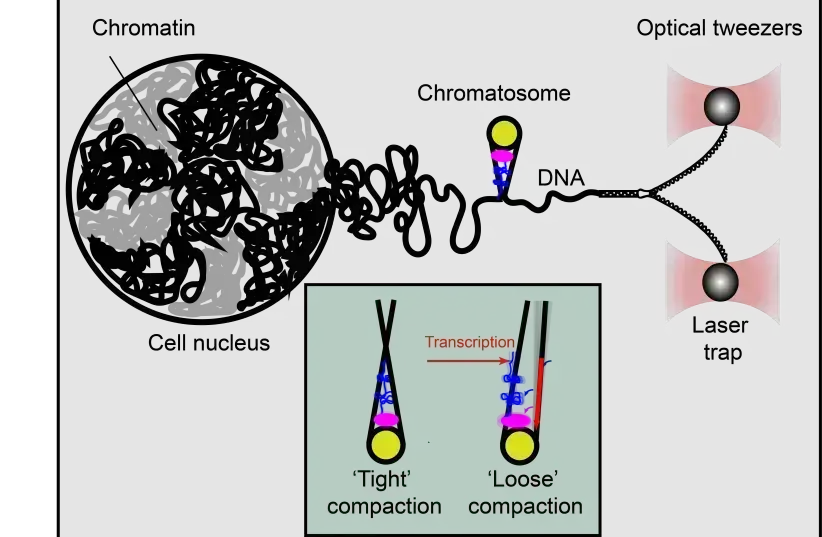
With this laser tweezer, Rudnizky, under the supervision of Profs. Ariel Kaplan and Philippa Melamed were able to slowly detach one strand of DNA from the rest of the strands. The process functions in a manner similar to unzipping a zipper, slowly removing the strand from the chromatosome. It isn’t a smooth process though, as the DNA strand can get stuck if it makes even the slightest contact with a histone. When that happens, more force is applied to advance further.
And as it turns out, histone-DNA contact is far more extensive, with chromatosomes being much larger than was previously believed. The linker histones themselves were also surprisingly flexible in structure because there were two different shapes the chromatosome can shift between, a symmetric and compact one and a relaxed and asymmetric one.
But it is possible to externally control the transition between these shapes through transcription mechanisms in the cells.
As suggested in the findings, published in the academic journal Molecular Cell, it is possible that the cell utilizes this transition to regulate its access to the DNA.
These findings are extremely significant, as they shed light on poorly understood functions of genome expression, which can further knowledge on the role of chromatin and chromatosomes in health and diseases.
HAIFA RESEARCHERS USE LASER ‘TWEEZERS’ TO STUDY STRUCTURE AND DYNAMICS OF DNA PACKAGING IN ALL CELLS
Bind them as a sign on your hand and let them serve as a symbol on your forehead.
Deuteronomy 6:8 (The Israel BibleTM)
Article published at www.israel365news.com on July 6, 2021.

Each one of the cells in our bodies contains DNA, which dictates the instructions needed for living things to develop and function. Incredibly, a total of two meters of DNA is packaged in each tiny cell’s nucleus, which is just tens of microns in size. This is made possible to packaging the DNA into a compact structure called chromatin.
The basic level of chromatin organization is provided by wrapping the DNA around proteins called histones in a spool-like structure that resembles beads on a string. Then, more complex structures called chromatosomes are formed with the help of a special histone known as a linker histone that connects the strings.
But scientists have not known much about the structure and dynamics of chromatosomes, leaving the most basic questions of how they bind DNA.
Now, researchers in the Biology Faculty of the Technion-Israel Institute of Technology in Haifa have used “laser tweezers” to accomplish this. Their study, just published in the journal Molecular Cell under the title “Extended and dynamic linker histone-DNA Interactions control chromatosome compaction,” was conducted by Dr. Sergei Rudnizky under the supervision of Profs. Ariel Kaplan and hilippa Melamed.
Packaging of the genome is essential for it to fit into the cell, but it also reduces the accessibility to the cellular machines that read the DNA and transcribe the genes. Therefore, the distinct packaging at a particular gene will have a huge impact on its expression in ways that are only beginning to be unraveled.
In particular, linker histones are known to play a key role in this organization of the genome – and their malfunction can lead to serious diseases including cancer and autism.
The lack of understanding of these crucial processes stems from the dynamic nature of linker histones, which makes it challenging to investigate them using conventional methods based on sampling a huge number of molecules simultaneously.
Kaplan’s lab developed a unique method based on “optical tweezers” – an approach that allows researchers to capture individual chromatin molecules and exert forces on them with the help of a focused laser beam.
In these experiments, one strand of DNA is slowly detached from its complementary strand in a manner similar to a zipper being unzipped, through the entire structure of a chromatosome. The principle of the measurement is simple – at points where a histone makes contact with the DNA, even in the weakest way, the zipper gets stuck, and more force needs to be applied to overcome the histone-DNA contact and advance into the structure.
Using this approach, the team discovered that contacts between histones and DNA are far more extensive than previously known and that chromatosomes are, in fact, much larger than previously thought. They also they found a surprising flexibility in the structure of linker histones, as two different chromatosome shapes exist – one symmetric and compact and the second asymmetric and more relaxed.
Amazingly, transition between these shapes in an individual molecule can be externally controlled by the transcription machinery itself. This suggests that the cell uses the transition between stable and unstable forms of a chromatosome to regulate access to the DNA in a controlled manner. Given the key role played by chromatosomes in maintaining proper expression of our genome, these findings add an important layer to our understanding of the role of chromatin architecture in health and disease.
